This tutorial is based on the blog post in this webpage: https://www.bayesianpolitik.me/2018/10/16/analysis_hurricane.html. I strongly recommend you to read the blog post first to get an idea of the data and the context.
There is a GitHub repository for this tutorial if you want to run it by yourself available here: https://github.com/ian-flores/mortality_analysis_tutorial
DISCLAIMER: I’m trying this new thing of the tutorials for the analyses I will be making now on as blog posts. It would be nice/helpful if I could get feedback. All kinds of feedbacks are welcomed. Feel free to use the Contact tab to contact me.
Notebook Content:
- #1) Getting and Cleaning Census Data
- How to get data from the census
- How to extract and manipulate data coming from a single table
- #2) Getting and Cleaning Mortality Data
- Cleaning data
- Dealing with strings in Spanish
- Grouping, summarizing, and splitting by different variables
- #3) Mapping Mortality Rates
- Creating maps in Python
- Joining data from different sources
- Dealing with spatial data in Python
- #4) Estimating Mortality Rates in Puerto Rico
- Bayesian Estimation with
PyMC3 - How to draw conclusions from data
- Bayesian Estimation with
Census Data¶
The United States established in 1840 a central office to oversee the census of the population of its states and territories. This office today is known as the United States Census Bureau. They perform a decenial census, the last one being in 2010. With this information they produce what is called PEPANNRES - Annual Estimates of the Resident Population in which they do population estimates by different geographic levels. In March 2018 they published the estimates for municipalities in the year 2017. This is exactly what we need for our analysis.
Getting the Data¶
The Census Bureau makes their data available through many pages and API's. Here we are going to use the FactFinder webpage and download the data manually, but it is worth noting that there are API wrappers in Python such as:
- Census - https://github.com/datamade/census
- Census Areas - https://github.com/datamade/census_area
If you want to get the data manually it is available in the following LINK. However, it is included as a zip file in the repository for reproducibility purposes.
Let's code!¶
Load the libraries¶
- pandas is a library to do data wrangling and dealing with data in general.
- os is a library that allows us to interact with the computer in a lower level than python.
- unidecode is a library that helps us deal with the names of the municipalities in Spanish. (Remember, PR is a Spanish speaking nation :D)
import pandas as pd
import os
import unidecode
Extract the data¶
The data is stored in a compressed form, we need to extract it and save it in a place where we can use it.
Steps:
- Extract the data ----
unzipcommand - Store it the
data/folder -----d data/command
os.system("unzip ../data/PEP_2017_PEPANNRES.zip -d ../data/")
Load the data¶
pd.read_csv is a function from the pandas package that allows us to load Comma Separated Values (CSV) files into Python with ease. The head() method let's us see the top rows of the data frame.
data = pd.read_csv("../data/PEP_2017_PEPANNRES.csv", encoding='latin-1')
data.head()
Dealing with names in Spanish¶
Spanish is a beautiful language, however, when dealing with names when programming, sometimes it's hard given some inconsistencies. This comes because of a wrong encoding of the files, typing some of the names in English and some in Spanish, or simply skiping 'weird' characters. To deal with this inconsistencies, we convert the names to their english-letter representation and to an upper case. This way we standarize all the names for all the files.
Steps:
- Extract the names of the municipalities from the strings
- Convert the names to english-letter representation
- Convert the strings to upper case format
municipalities = []
for i in range(0, 78):
muni_raw = data['GEO.display-label'][i].split(" Municipio, Puerto Rico")[0]
muni_clean = unidecode.unidecode(muni_raw)
muni_upper = muni_clean.upper()
municipalities.append(muni_upper)
data['ResidencePlace'] = municipalities
data_processed = data[['ResidencePlace', 'respop72017']]
data_processed.head()
Save the progress | Clean our directories¶
Let's save the population estimates for 2017 to use the file later in our pipeline
data_processed.to_csv("../data/census.csv")
Finally, clean the data directory to only remain with the files we need to reproduce this analysis and produce it's output.
os.system("rm ../data/PEP_2017_PEPANNRES.csv ../data/PEP_2017_PEPANNRES_metadata.csv ../data/PEP_2017_PEPANNRES.txt ../data/aff_download_readme.txt")
Optional Questions¶
1) What is the municipality with the highest population? How about the least?
2) Which municipality changed the most between 2016 and 2017?
3) How about between 2010 and 2017?
4) Can you find the municipality that has stayed the most stable (less variability) between all these years?
5) Are there some municipalities consistingly losing people and other winning them?

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Purpose of this Notebook¶
Mortality Data¶

Hurricane Maria struck Puerto Rico the 20th of September of 2017 at 6:15 a.m. entering through the municipality of Yabucoa as a Category 4 hurricane. According to the governments of Puerto Rico and the U.S. Virgin islands, the cost of the damage is estimated in $102 billion USD1. However, the impact wasn't only economical. Following a lawsuit presented by the Center for Investigative Journalism (CPI, by its spanish initials) and CNN, the Government of Puerto Rico, and more specifically the Demographic Registry, was forced to publish the individual-level data of all the deaths occurred from September 20, 2017 to June 11, 2018. This data is available in the following Dropbox Link from the CPI.
Let's code!¶
Load the libraries¶
import pandas as pd
import os
Here we are downloading the data directly from Dropbox and storing it under the data/ folder in the mortality.xlsx file.
os.system("wget --output-document='../data/mortality.xlsx' 'https://www.dropbox.com/s/k4wrb1ztwu0fwxh/Base%20de%20Datos%20Mortalidad%20en%20PR%20de%20septiembre%2018%20de%202017%20a%2011%20de%20junio%20de%202018%20entregada%20por%20Gobierno%20de%20PR%20al%20CPI.xls?dl=0'")
data = pd.read_excel("../data/mortality.xlsx")
data.head()
Filtering the data¶
- We are using the ResidencePlace column as the column for the Municipality
- There is people that died in Puerto Rico, but there residence place is not Puerto Rico, we want to exclude those data points from the analysis.
data[data.ResidencePlace.str.contains('PUERTO RICO')]
- We also want to exclude those people we don't know their municipality of residence.
data[data.ResidenceZone != 'DESCONOCIDO']
data = data[data.ResidencePlace.str.contains('PUERTO RICO')]
data = data[data.ResidencePlace != 'PUERTO RICO, DESCONOCIDO']
data = data[data.ResidenceZone != 'DESCONOCIDO']
Grouping the data¶
We want to compute the number of deaths in each zone of each municipality:
- Group by
ResidencePlaceandResidenceZonedata.groupby(['ResidencePlace', 'ResidenceZone'])
- Calculate the number of deaths per zone of each municipality
data.groupby(['ResidencePlace', 'ResidenceZone']).size().reset_index(name='Deaths')
df_grp = data.groupby(['ResidencePlace', 'ResidenceZone']).size().reset_index(name='Deaths')
df_grp.head()
Extract the name of the municipalities from the ResidencePlace column¶
municipalities = df_grp.ResidencePlace.tolist()
for i in range(0,len(municipalities)):
municipalities[i] = municipalities[i].split("PUERTO RICO, ")[1]
df_grp.ResidencePlace = pd.Series(municipalities)
df_grp.head()
Save our DataFrame as a CSV for future analysis¶
df_grp.to_csv("../data/mortality_grouped.csv")
Optional Questions¶
1) Did more women died or men?
2) Which month had the higher mortality?
3) Which was the average age of death? Did more infants died or older people?
4) Do you see any patterns in years of education?
5) Do you notice any patter in the ResidencePlace vs the Death Registry municipality? Why is this?

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Let's code¶
Load the libraries¶
- Here we see the
%matplotlib inlinecommand, this tells the matplotlib package to display the plots in this notebook. - We are seeing as well the
geopandaslibrary for the first time. This is a very useful library to handle spatial data in Python.
%matplotlib inline
import matplotlib.pyplot as plt
import geopandas as gpd
import pandas as pd
import numpy as np
import unidecode
import os
Get the data¶
- We are downloading a GeoJSON file, which is a text file formatted to contain spatial information by hierarchies.
- Hierarchies can vary but it goes from the world, to a region, to a country, to a municipality, etc.
- This GeoJSON is available from Miguel Rios GitHub repository.
os.system("wget 'https://raw.githubusercontent.com/miguelrios/atlaspr/master/geotiles/pueblos.json'")
os.system("mv 'pueblos.json' '../data/'")
geo_muni = gpd.read_file('../data/pueblos.json')
geo_muni.head()
On the geometry column we see a POLYGON object which is a spatial object that defines the boundary coordinates of the municipalities.
Plot the municipalities¶
- Calling the GeoDataFrame by its name with the
.plot()object, will plot the geometry column which in this case is our polygons of the municipalities
geo_muni.plot()
Cleaning the municipalities name¶
- This is the same procedure as in notebook_01.
municipalities = []
for i in range(0, 78):
muni_raw = geo_muni['NAME'][i].split(" Municipio, Puerto Rico")[0]
muni_clean = unidecode.unidecode(muni_raw)
muni_upper = muni_clean.upper()
municipalities.append(muni_upper)
geo_muni['NAME'] = municipalities
Loading Census Data¶
census_data = pd.read_csv("../data/census.csv")
census_data = census_data.drop(['Unnamed: 0'], axis = 1)
census_data.head()
Loading Mortality Data¶
mortality_data = pd.read_csv("../data/mortality_grouped.csv")
mortality_data = mortality_data.drop(['Unnamed: 0'], axis = 1)
mortality_data.head()
Calculating mortality rates¶
- We first join the census data to the mortality DataFrame. This order matters as the mortality DataFrame has more information than the census data.
- We then divide the number of deaths by the number of people in each municipality and the multiply by 1000 to get the mortality rate per 1000 people.
df = mortality_data.merge(census_data, on='ResidencePlace')
df['death_rate'] = (df['Deaths']/df['respop72017']) * 1000
df.head()
Join the mortality data and the spatial data¶
geo_muni['death_rate'] = list(df.groupby(['ResidencePlace'])['death_rate'].sum())
Let's plot the death rate by municipality and compare it with the trajectory of the hurricane¶
figure, ax = plt.subplots(1)
geo_muni.plot(column = 'death_rate',
scheme = 'quantiles',
legend = True,
ax = ax,
edgecolor='1',
linewidth = 0.3)
fig = plt.gcf()
fig.set_size_inches(7, 6)
fig.set_dpi(125)
plt.title("Mortality rate per 1,000 individuals\n Timeframe from September 20, 2017 to June 2018")
ax.set_axis_off()
plt.axis('equal')
plt.show()
 Source: NYTimes
Source: NYTimes
Optional Questions¶
1) Do you see a pattern between the trajectory of the Hurricane and the mortality rates' spatial distribution?
2) Do you see a difference between rural zones and urban zones? Their means? Their variances? Maybe remember the groupby function to do this step. We will go way more in depth in the next notebook.

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Purpose of this Notebook¶
Learning¶
- Getting to know the
PyMC3library - Starting to think about Bayesian methods
- Think about uncertainty in the estimates instead of significance vs no-significance (*Not a fan of the p-value)
Project¶
- Estimate the mortality rate for urban areas as compared to rural areas.
- Identify which area needs more attention for future disasters.
Bayesian methods¶
Bayesian Paradigm¶
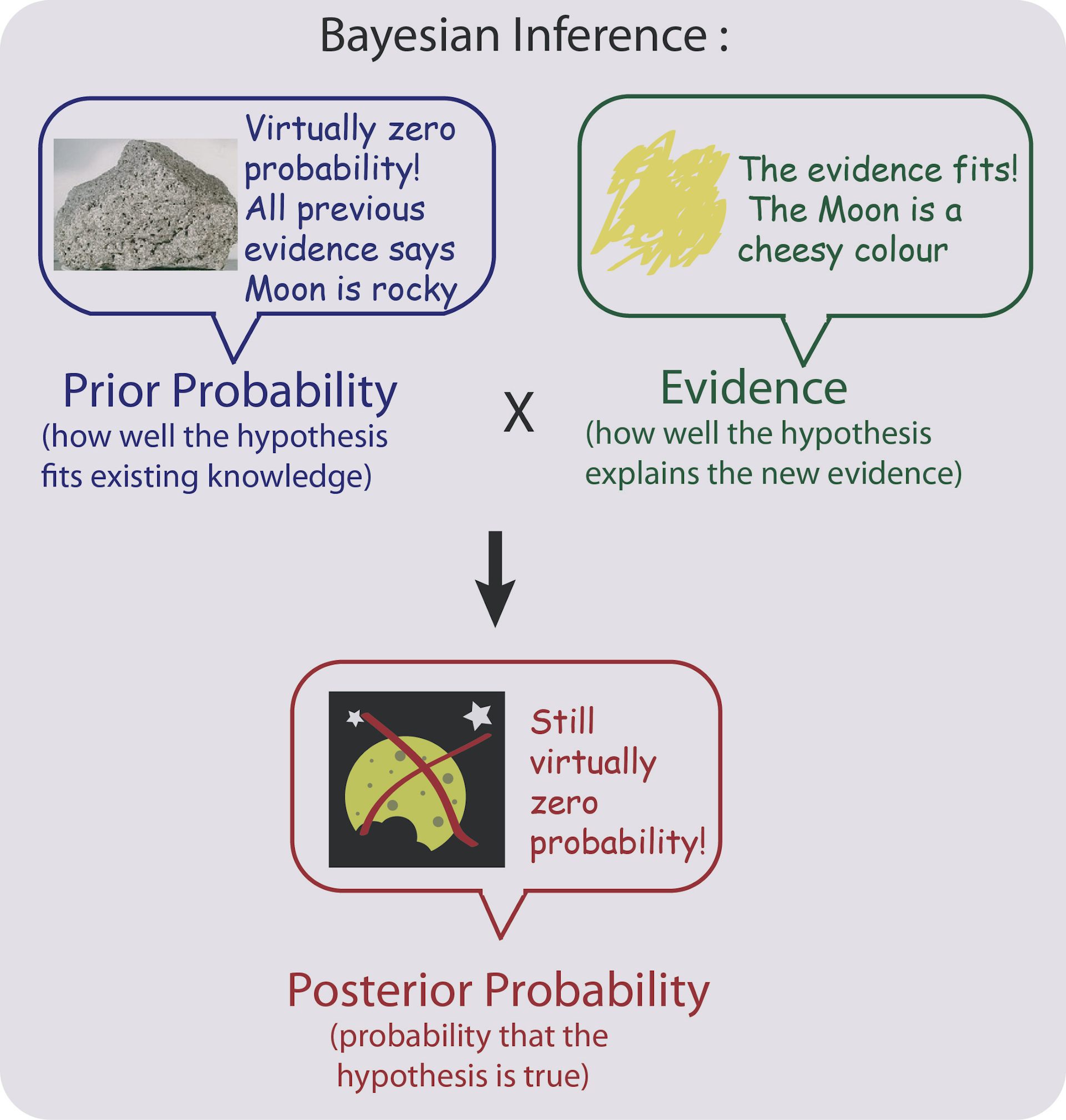
What if we have previous knowledge, or have expert knowledge regarding a certain subject that we would like to incorporate? With Bayesian methods, we can set something that is called a prior, which is a distribution for each parameter of the phenomenom we want to estimate or test. We combine this priors with the data (a.k.a Likelihood) to obtain a posterior distribution which allows us the necessary inference.
DISCLAIMER: Priors are controversial as some people have suggested that one can skew the results of the posterior distribution by setting the priors to certain values. This might be true in cases of few data points, however, in an open and reproducible science approach this should not be a concern as everyone alse can see your work. This is a big difference between using tools such as
RandPythonas compared to other click-and-touch software.
History¶
- Thomas Bayes discovered it.
- Pierre-Simon Laplace greatly expanded the methods working with demographic data from France
- Didn't know about Bayes discovery as British-French relations at the moment weren't great, even though Laplace was almost two centuries later.
- Also invented the Central Limit Theorem, a crucial mathematical concept to understand computation bayesian methods.
- Invented the mathematical system for inductive reasoning in probability.
Uses¶
- Misile production estimation
- Estimating the number of tanks the German Nazis were producing per factory
- Cracking the encryption
- Breaking the Lorenz cipher using Bayesian methods
- Coast Guard Searchs
- Searhing by quadrants for submarines in the Atlantic Ocean
- Election Prediction
import pandas as pd
import numpy as np
import pymc3 as pm
import matplotlib.pyplot as plt
Load the data¶
census_data = pd.read_csv("../data/census.csv")
census_data = census_data.drop(['Unnamed: 0'], axis = 1)
mortality_data = pd.read_csv("../data/mortality_grouped.csv")
mortality_data = mortality_data.drop(['Unnamed: 0'], axis = 1)
Calculating mortality rates¶
- We first join the census data to the mortality DataFrame. This order matters as the mortality DataFrame has more information than the census data.
- We then divide the number of deaths by the number of people in each municipality and the multiply by 1000 to get the mortality rate per 1000 people.
df = mortality_data.merge(census_data, on='ResidencePlace')
df['death_rate'] = (df['Deaths']/df['respop72017']) * 1000
df.head()
Here we are making two DataFrames, one with the information for the rural zones and one for the urban zones.
rural = df[df['ResidenceZone'] == 'RURAL']['death_rate']
urban = df[df['ResidenceZone'] == 'URBANO']['death_rate']
PyMC3, the package we are using for Bayesian estimation, requires the data in a certain structure, which is what we are doing in this code cell below.
total = pd.DataFrame(dict(death_rate = np.r_[rural, urban],
group = np.r_[['rural']* len(rural),
['urban'] * len(urban)]))
total.head()
Priors¶
As mentioned before, when doing bayesian estimation we need to set our priors. In this case I'm stating that our priors for the mean mortality rate in each zone should be close to the total death rate. In this case, I am modeling this priors with a TruncatedNormal distribution, bounded between 0 and a 1000, from the PyMC3 library. This would mean the extreme of either no one dying or everyone dying respectively. As we don't know much about the variability of the data, we model it for both zones with a Uniform distribution between 0 and 10. v is the degrees of freedom, we are centering the mean around 2 in this case with a Gamma distribution.
mu_total = total.death_rate.mean()
std_total = total.death_rate.std()
with pm.Model() as model:
rural_mean = pm.TruncatedNormal('rural_mean',
mu = mu_total,
sd = std_total,
lower = 0, upper = 1000)
urban_mean = pm.TruncatedNormal('urban_mean',
mu = mu_total,
sd = std_total,
lower = 0, upper = 1000)
std_low = 0
std_high = 10
with model:
rural_std = pm.Uniform('rural_std', lower = std_low,
upper = std_high)
urban_std = pm.Uniform('urban_std', lower = std_low,
upper = std_high)
with model:
v = pm.Gamma('v', alpha = 3, beta = 1)
Likelihood¶
In this code cell we are describing the final distribution, which is a StudentT distribution to compare the mean of the two groups at the end. We are passing the different arguments to the distribution and at the end with the observed argument we are feeding the model the data we have observed.
with model:
lambda_rural = rural_std**-2
lambda_urban = urban_std**-2
rural_try = pm.StudentT('rural', nu = v, mu = rural_mean,
lam = lambda_rural, observed = rural)
urban_try = pm.StudentT('urban', nu = v, mu = urban_mean,
lam = lambda_urban, observed = urban)
Here, we are calculating the differences of the resulting distributions to see if there is a difference between both zones.
with model:
diff_of_means = pm.Deterministic('difference of means',
rural_mean - urban_mean)
diff_of_stds = pm.Deterministic('difference of stds',
rural_std - urban_std)
Sampling¶
We are going to take samples from this distributions, in our case it will be 20,000 samples, over 4 cores, discarding the first 15,000 samples in each chain. This leaves us with a total of 140,000 samples from the distributions.
with model:
trace = pm.sample(20000, cores = 4, tune = 15000)
Let's look at the posterior of the mean of the mortality rate in urban zones. What do you see?
pm.plot_posterior(trace, varnames = ['urban_mean'], color = '#0d98ba')
We can observe that the mean is centered around 2.142 with a 95% chance that the real number is between 1.862 and 2.429, which means that we can say that in average, for every 1000 people, 2 died in urban zones. Below we can see that the standard deviation for the urban zones is 1.063.
pm.plot_posterior(trace, varnames = ['urban_std'], color = '#0d98ba')
Now, let's look at the posterior of the mean of the mortality rate in rural zones. Do you notice a difference?
pm.plot_posterior(trace, varnames = ['rural_mean'], color = '#0d98ba')
We can observe that the mean is centered around 4.126 with a 95% chance that the real number is between 3.81 and 4.451, which means that we can say that in average, for every 1000 people, 4 died in rural zones. Below we can see that the standard deviation for the rural zones is 1.207, which means that the variability between urban and rural zones was similar.
pm.plot_posterior(trace, varnames = ['rural_std'], color = '#0d98ba')
However, how does the rural distribution as a whole compare to the urban distribution?
pm.plot_posterior(trace, varnames = ['difference of means'], color = '#0d98ba', ref_val=0)
Above we can see that approximately, in avergae, 2 more people died in rural zones as compared to urban zones.
pm.forestplot(trace, varnames = ['rural_mean', 'urban_mean'])
Finally, if we compare the different chains of the sample visually we can see that they are all very diiferent between zones.
Optional Questions¶
1) Do you think this analysis could incorporate other variables?
2) How do you think the results compare to the frequentist paradigm? Do you prefer to know the uncertainty in your results or have a single number to which you can refer to?
3) I encourage you to play with the PyMC3 package more as you can do very exciting things. (Such as model the result of football games)
References¶
This work was inpired in part by this work done by the PyMC3 development team:https://docs.pymc.io/notebooks/BEST.html

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.